The key difference between BLAST and FastA is that the BLAST is a basic alignment tool available at National Center for Biotechnology Information website while FastA is a similarity searching tool available at European Bioinformatics Institute website.
BLAST and FastA are two software that is widely in use to compare biological sequences of DNA, amino acids, proteins, and nucleotides of different species and look for their similarities. These algorithms were written keeping speed in mind. Because, when scientists were able to isolate DNA in the laboratory in the mid-1980s, it raised a need to compare and find identical genes for further research at high speed. Hence, these two softwares were developed in a way that user can do a fast search of similar sequences with that of their query sequences.
BLAST is an acronym for Basic Local Alignment Search Tool and uses the localized approach in comparing the two sequences. FastA is a software referring to Fast A where A stands for All. Here, the software works with alphabets like Fast A for DNA sequencing and Fast P for protein. Both BLAST and FastA are very fast in comparing any genome database and are, therefore, very viable monetarily as well as in saving time.
CONTENTS
1. Overview and Key Difference
2. What is BLAST
3. What is FastA
4. Similarities Between BLAST and FastA
5. Side by Side Comparison – BLAST vs FastA in Tabular Form
6. Summary
What is BLAST?
BLAST is one of the most widely-used bioinformatics software that was developed in 1990. Since then, it is available to everyone at the NCBI site. Furthermore, any person can access this software and use it. Moreover, BLAST is a software that needs input data or sequences in FastA format. But, it gives output data in plain text, HTML, or XML format. BLAST works on a principle of searching for localized similarities between two sequences and short list the similar sequences, and after that, it searches for neighbourhood similarities.
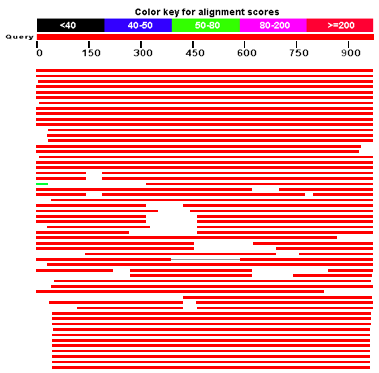
Figure 01: BLAST Results
Thus, this software searches for a high number of similar local regions and gives the result on reaching a threshold value. However, this process differs from the earlier software,which searches the entire sequence first and then does the comparison, and hence, took a lot of time.
Apart from the above similarity checking, BLAST has many other uses such as the DNA mapping, comparing two identical genes in different species, creating a phylogenetic tree, etc.
What is FastA?
FastA is a protein sequence alignment software. David J. Lipman and William R. Pearson described this software in 1985. Though the initial use of this software was to compare the protein sequences only, the modified version of it was able to compare DNA sequences as well. Here, this software uses the principle of finding the similarity between two sequences statistically. It matches one sequence of the DNA or protein with another sequence by local sequence alignment method.
Figure 02: FastA
However, it searches for local regions for similarity, but not the best match between two sequences. Since this software compares localized similarities at times, it can come up with mismatches as well. In a sequence, FastA takes a small part known as k-tuples, where the tuples can be from 1 to 6 and matches with the k-tuples of the other sequence. At the end of the matching process, when it reaches a threshold value, it produces the result.
What are the Similarities Between BLAST and FastA?
- BLAST and FastA are bioinformatics tools used to compare protein and DNA sequences for similarities.
- Also, both programs use a scoring strategy to make comparisons between the sequences.
- Furthermore, both tools produce highly accurate results.
What is the Difference Between BLAST and FastA?
BLAST is a tool to check the similarity between biological sequences. On the other hand, FastA is another program that facilitates the similarity checking of protein and DNA sequences. However, in comparison to FastA, BLAST software is very popular since it produces more accurate and fast results. Hence, this is the difference between BLAST and FastA. Furthermore, unlike FastA, the BLAST program is modifiable according to the need of the user. Therefore, this is another difference between BLAST and FastA.
The below infographic on difference between BLAST and FastA provides more details.

Summary – BLAST vs FastA
BLAST and FastA are two programs that allow the user to compare his query sequence with the sequences in the existing databases and check the similarities. The initial purpose of FastA was to compare only protein sequences. But, the modified version of this software facilitates both protein and DNA sequence comparisons. Though FastA is good software, most people use BLAST alignment tool since it is more popular and produces more accurate and fast results than FastA. Also, BLAST tool is modifiable according to the user requirements. In brief, this summarizes the difference between BLAST and FastA.
Reference:
1.Madden, Thomas. “The BLAST Sequence Analysis Tool.” Current Neurology and Neuroscience Reports., U.S. National Library of Medicine, 15 Mar. 2013. Available here
Image Courtesy:
1.”CCDC132 Blast Results”By NCBI Blast – NCBI Blast, (Public Domain) via Commons Wikimedia
2.”FAM149A Promoter region (FASTA format)”By LarsonGCD – Own work, (CC BY-SA 3.0) via Commons Wikimedia
ncG1vNJzZmivp6x7pbXFn5yrnZ6YsqOx07CcnqZemLyue8OinZ%2Bdopq7pLGMm5ytr5Wau26uy5qqrWWRo7FuwtJmnZqrpJZ8